melanoma
melanoma.Rmd
library(survival)
library(dplyr)
library(ISwR)
library(devtools)
library(survRM2)
# library(smim)
load_all()Data Cleanning
- Pattern
- 1: observed event time
- 2: MAR imputation
- 3: MNAR imputation
- Delta
- 1: no adjustment
- larger than 1: hazard is larger than MAR
- less than 1: hazard is smaller than MAR
data("melanom")
db <- melanom
db <- db %>% mutate(
time = days/30.25, # unit in Month
status = ifelse(status %in% c(1,3), 0, 1 ),
group = ifelse(thick > median(thick), 1, 0),
pattern = ifelse(status == 1, 1, ifelse(time < 8, 2, 3)),
log_thick = log(thick)
) %>% arrange(group, status, pattern, time) %>%
mutate(id = 1:n())
time <- db$time
status <- db$status
group <- db$group
pattern <- db$pattern
x <- db %>% select(sex, ulc)
tau <- 10*12
# db0 <- db %>% select(time, status, sex, log_thick)
# fit <- coxph(Surv(time, status) ~ rep(1, nrow(db0)), data = db0, x = TRUE, y = TRUE)MAR analysis
rmst2(time, status, group, tau= tau)
#>
#> The truncation time: tau = 120 was specified.
#>
#> Restricted Mean Survival Time (RMST) by arm
#> Est. se lower .95 upper .95
#> RMST (arm=1) 92.215 3.428 85.496 98.934
#> RMST (arm=0) 83.333 2.665 78.110 88.555
#>
#>
#> Restricted Mean Time Lost (RMTL) by arm
#> Est. se lower .95 upper .95
#> RMTL (arm=1) 27.785 3.428 21.066 34.504
#> RMTL (arm=0) 36.667 2.665 31.445 41.890
#>
#>
#> Between-group contrast
#> Est. lower .95 upper .95 p
#> RMST (arm=1)-(arm=0) 8.882 0.372 17.393 0.041
#> RMST (arm=1)/(arm=0) 1.107 1.005 1.218 0.039
#> RMTL (arm=1)/(arm=0) 0.758 0.572 1.003 0.053
fit <- survfit(Surv(time, status) ~ group, data = db)
# Visualize with survminer
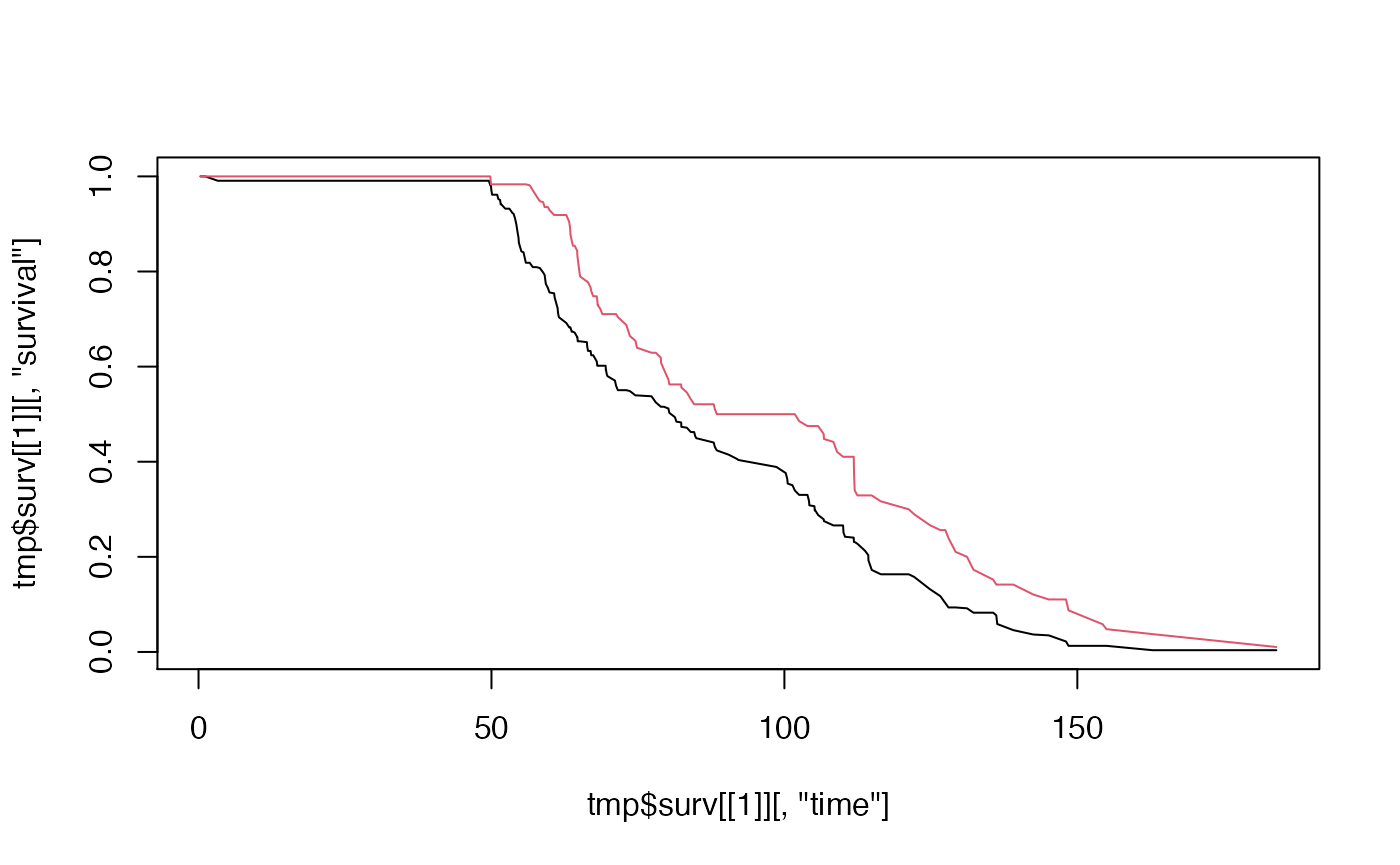
plot(fit, col = c(1,2))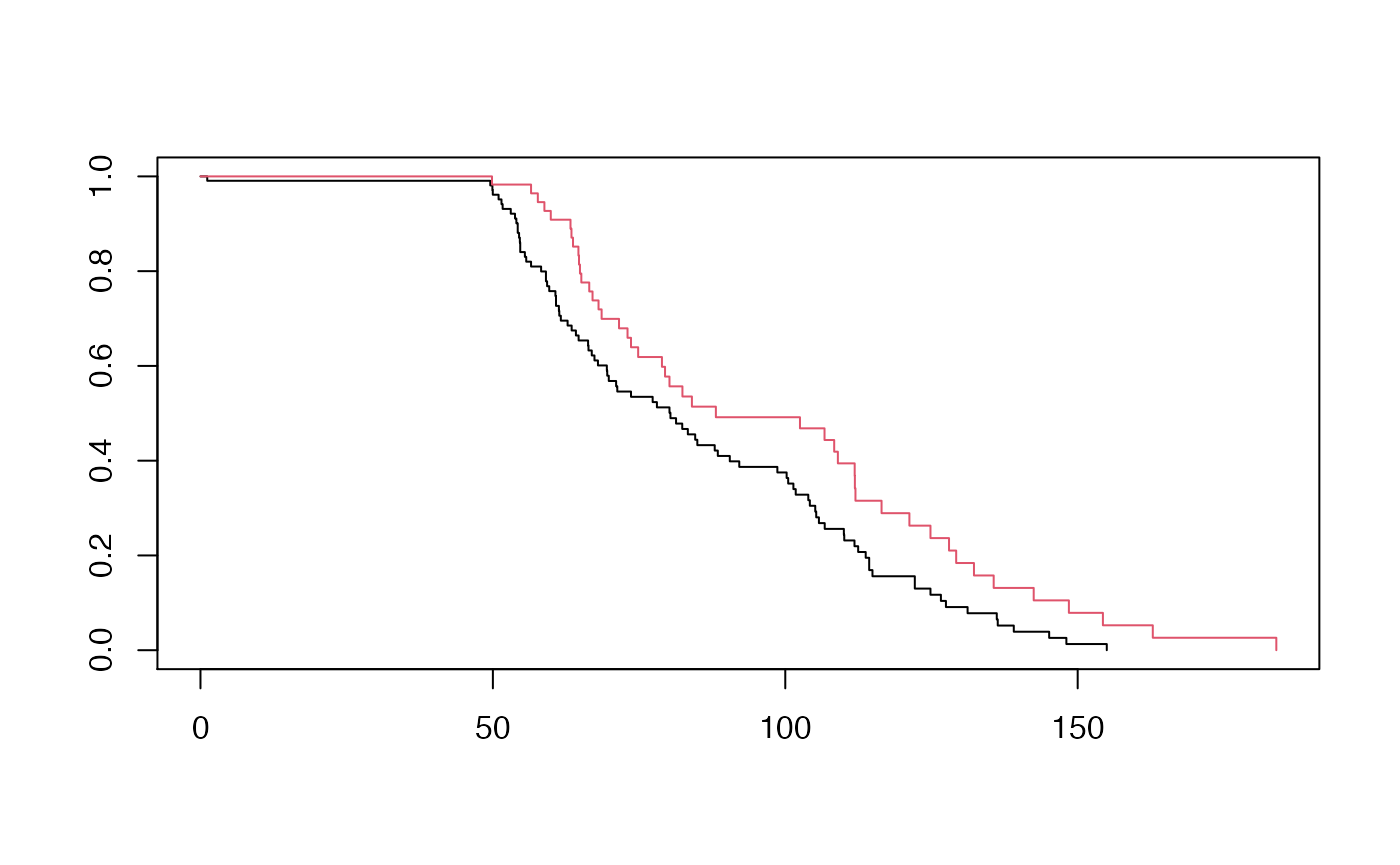
Delta Adjusted Imputation (MAR)
RMST within group
delta <- c(1,1,1)[pattern] # the third number control delta adjustment for MNAR
tmp <- rmst_delta(time, status, x = rep(1, length(time)), group, pattern, delta, tau, n_mi = 5, n_b = 100, seed = 1234)
tmp$rmst
#> group rmst sd wb_sd
#> [1,] 0 83.55763 2.664831 2.190611
#> [2,] 1 92.64380 3.307655 1.806165RMST between group
diff_rmst <- function(rmst, sd){
diff <- diff(rmst)
diff_sd <- sqrt( sum(sd^2) )
p_val <- 2* (1 - pnorm( abs(diff/diff_sd) ))
c(diff = diff, sd = diff_sd, p = p_val)
}
rbind( diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "sd"]),
diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "wb_sd"]))
#> diff sd p
#> [1,] 9.08617 4.247577 0.032424164
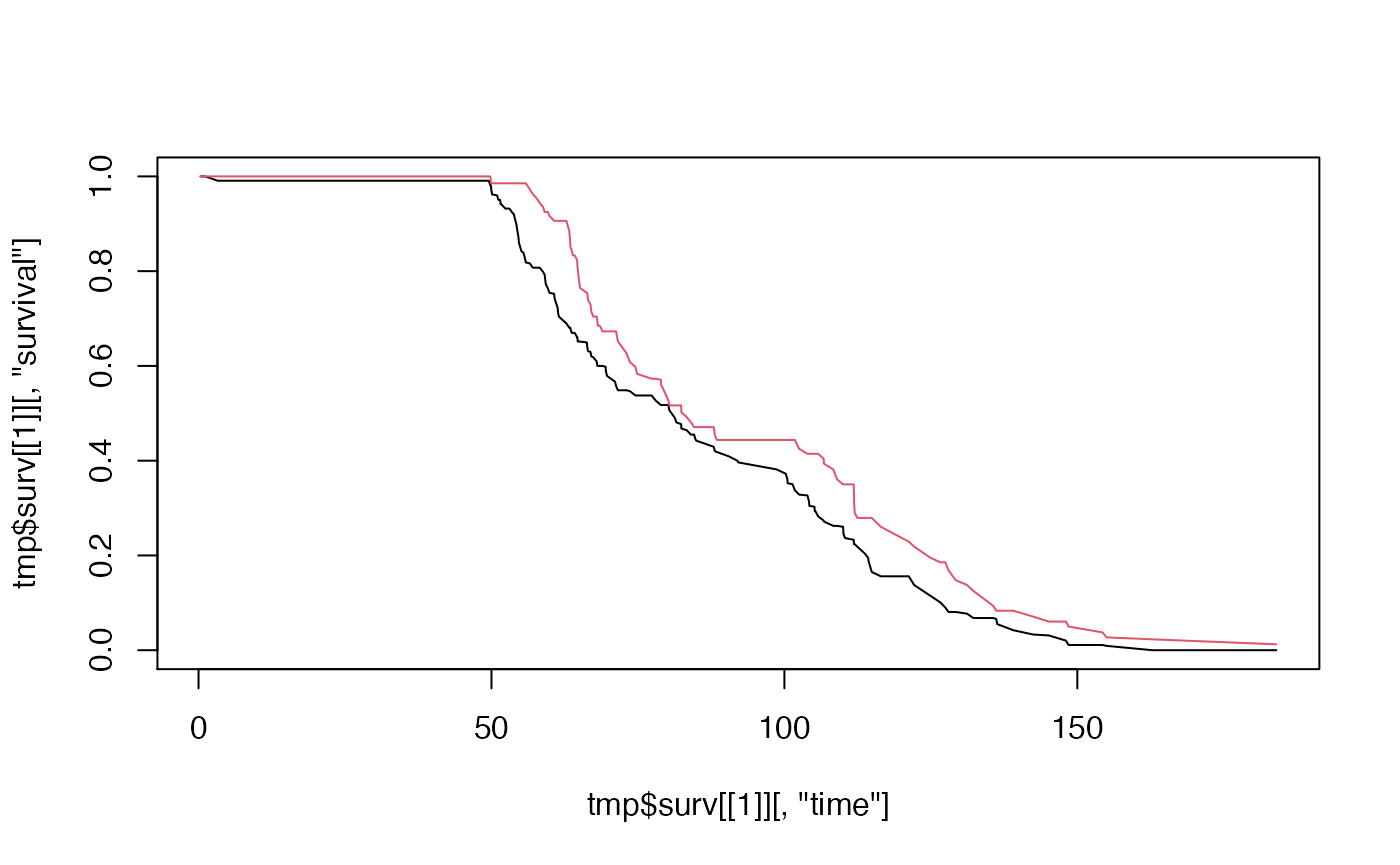
#> [2,] 9.08617 2.839192 0.001373007Delta Adjusted Imputation (MNAR)
RMST within group
delta <- c(1,1,1.25)[pattern] # the third number control delta adjustment for MNAR
tmp <- rmst_delta(time, status, x = x, group, pattern, delta, tau, n_mi = 5, n_b = 100, seed = 1234)
tmp$rmst
#> group rmst sd wb_sd
#> [1,] 0 83.27594 2.681406 2.210220
#> [2,] 1 89.57342 3.267553 1.936776RMST between group
diff_rmst <- function(rmst, sd){
diff <- diff(rmst)
diff_sd <- sqrt( sum(sd^2) )
p_val <- 2* (1 - pnorm( abs(diff/diff_sd) ))
c(diff = diff, sd = diff_sd, p = p_val)
}
rbind( diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "sd"]),
diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "wb_sd"]))
#> diff sd p
#> [1,] 6.297474 4.226919 0.13626375
#> [2,] 6.297474 2.938737 0.03211963