Example: actg175 dataset
actg175.Rmd
library(survival)
library(dplyr)
library(speff2trial)
library(devtools)
library(survRM2)
library(purrr)
# library(smim)
load_all()Data Cleanning
The dataset is avaiable in speff2trial::ACTG175.
# Load dataset
data(ACTG175)- Pattern
- 1: observed event time
- 2: MAR imputation
- 3: MNAR imputation
- Delta
- 1: no adjustment
- larger than 1: hazard is larger than MAR
- less than 1: hazard is smaller than MAR
# Analysis data: antiretroviral naive and no history of intravenous drug use
db <- ACTG175 %>% subset(drugs == 0 & strat == 1)
db <- db %>% mutate(time = days / 30.25,
age = (age - mean(age)) / sd(age),
status = cens,
group = arms,
pattern = ifelse(status == 1, 1, ifelse(time >= 24, 2, 3))) %>%
subset(group %in% c(0, 1)) %>%
mutate(group = ifelse(group == 0, 0, 1) )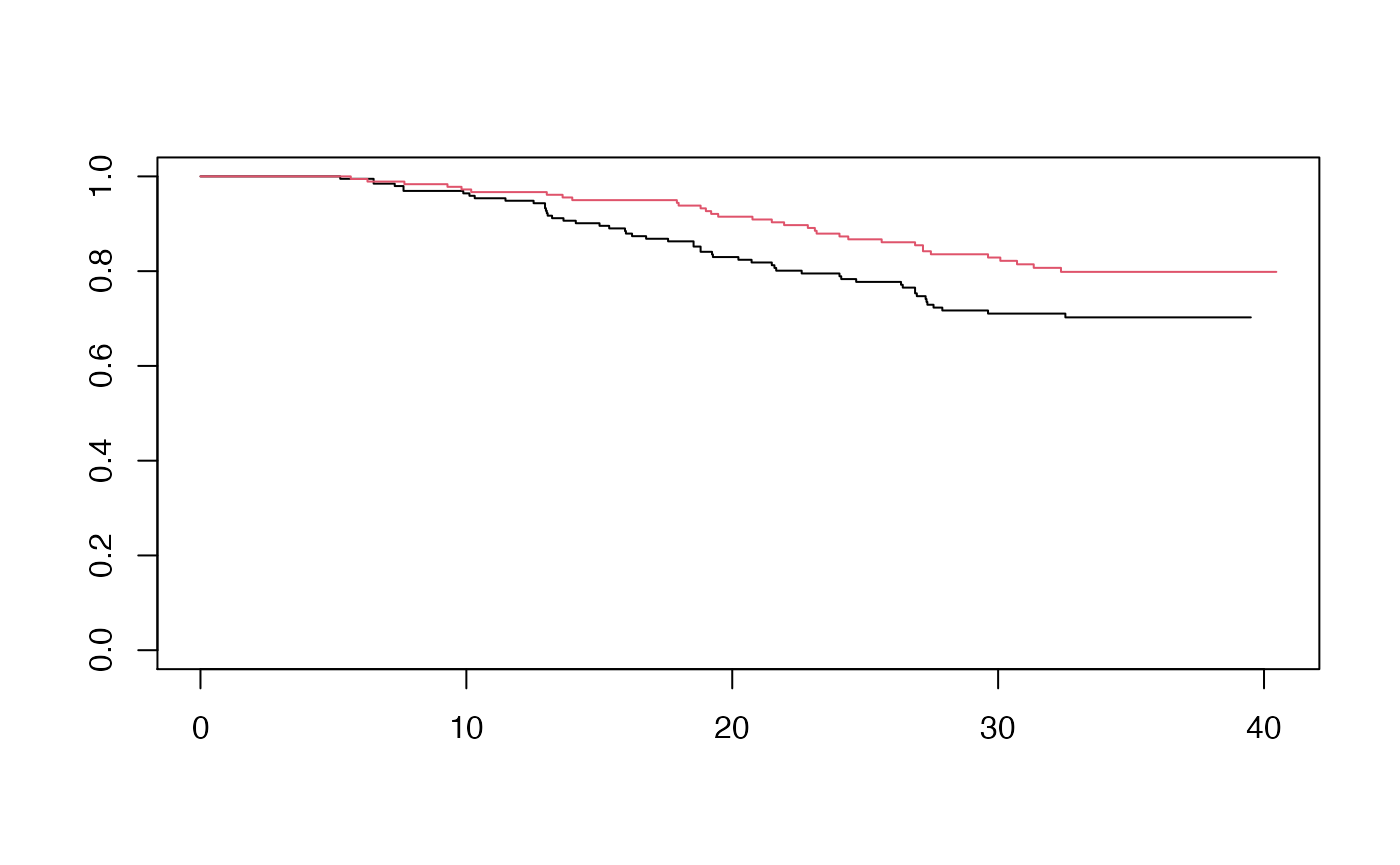
Summary statistics
nrow(db)
#> [1] 382
table(group)
#> group
#> 0 1
#> 197 185
table(status)
#> status
#> 0 1
#> 296 86
n_mi <- 50
n_b <- 100
seed <- 1234Delta Adjusted Imputation
RMST within group
.delta <- seq(1, 5, by = 0.5)
res <- map(.delta, function(delta0){
delta <- ifelse(group == 0, c(1,1,1)[pattern], c(1,1,delta0)[pattern]) # the third number control delta adjustment for MNAR
tmp <- rmst_delta(time, status, x = x, group, pattern, delta, tau, n_mi = n_mi, n_b = n_b, seed = seed)
tmp
})
res0 <- map(res, function(x) as.data.frame(x$rmst))
names(res0) <- .delta
res0 <- bind_rows(res0, .id = "delta")
res0 %>% mutate(
lower = rmst - 1.96 * sd,
upper = rmst + 1.96 * sd,
lower_wb = rmst - 1.96 * wb_sd,
upper_wb = rmst + 1.96 * wb_sd
) %>% mutate_if(is.numeric, formatC, digits = 2, format = "f")
#> delta group rmst sd wb_sd lower upper lower_wb upper_wb
#> 1 1 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 2 1 1.00 23.04 0.24 0.22 22.56 23.52 22.60 23.48
#> 3 1.5 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 4 1.5 1.00 23.02 0.25 0.22 22.54 23.50 22.58 23.46
#> 5 2 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 6 2 1.00 23.00 0.25 0.23 22.51 23.49 22.56 23.44
#> 7 2.5 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 8 2.5 1.00 22.98 0.25 0.23 22.49 23.47 22.54 23.43
#> 9 3 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 10 3 1.00 22.97 0.25 0.23 22.47 23.46 22.52 23.41
#> 11 3.5 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 12 3.5 1.00 22.95 0.25 0.23 22.45 23.45 22.51 23.40
#> 13 4 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 14 4 1.00 22.93 0.26 0.23 22.43 23.44 22.49 23.38
#> 15 4.5 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 16 4.5 1.00 22.92 0.26 0.23 22.42 23.42 22.47 23.37
#> 17 5 0.00 22.12 0.31 0.32 21.52 22.73 21.50 22.75
#> 18 5 1.00 22.90 0.26 0.23 22.40 23.41 22.45 23.35RMST between group
diff_rmst <- function(data){
rmst <- data$rmst
sd <- data$sd
diff <- diff(rmst)
diff_sd <- sqrt( sum(sd^2) )
p_val <- 2* (1 - pnorm( abs(diff/diff_sd) ))
print(c(diff = diff, sd = diff_sd, p = p_val))
return(1)
}
res0 %>% group_by(delta) %>% summarise(
diff_rmst = diff(rmst),
diff_sd = sqrt( sum(sd^2) ),
diff_wb_sd = sqrt(sum(wb_sd^2)),
lower = diff_rmst - 1.96 * diff_sd,
upper = diff_rmst + 1.96 * diff_sd,
p_val = 2* (1 - pnorm( abs(diff_rmst/diff_sd) )),
lower_wb = diff_rmst - 1.96 * diff_wb_sd,
upper_wb = diff_rmst + 1.96 * diff_wb_sd,
p_val_wb = 2* (1 - pnorm( abs(diff_rmst/diff_wb_sd) )),
) %>% mutate_if(is.numeric, formatC, digits = 3, format = "f") %>% data.frame()
#> delta diff_rmst diff_sd diff_wb_sd lower upper p_val lower_wb upper_wb
#> 1 1 0.915 0.394 0.389 0.143 1.686 0.020 0.151 1.678
#> 2 1.5 0.896 0.395 0.390 0.122 1.671 0.023 0.133 1.660
#> 3 2 0.877 0.397 0.390 0.100 1.655 0.027 0.113 1.642
#> 4 2.5 0.859 0.398 0.390 0.078 1.639 0.031 0.094 1.624
#> 5 3 0.843 0.399 0.391 0.062 1.625 0.034 0.078 1.609
#> 6 3.5 0.826 0.400 0.391 0.042 1.610 0.039 0.060 1.592
#> 7 4 0.810 0.401 0.391 0.024 1.596 0.043 0.043 1.577
#> 8 4.5 0.794 0.402 0.392 0.006 1.582 0.048 0.026 1.562
#> 9 5 0.778 0.403 0.392 -0.012 1.568 0.054 0.009 1.547
#> p_val_wb
#> 1 0.019
#> 2 0.021
#> 3 0.024
#> 4 0.028
#> 5 0.031
#> 6 0.035
#> 7 0.038
#> 8 0.043
#> 9 0.047Control Based Imputation
delta <- c(1,1,1)[pattern] # the third number control delta adjustment for MNAR
tmp <- rmst_control(time, status, x = x,
group = group,
ref_grp = 0, pattern = pattern, delta = delta, tau = tau,
n_mi = n_mi, n_b = n_b, seed = seed)
tmp$rmst
#> group rmst sd wb_sd
#> [1,] 0 22.12450 0.3091611 0.3183603
#> [2,] 1 23.00746 0.2478957 0.2247016RMST between group
- first row: Rubin’s rule
- second row: Wild bootstrap
diff_rmst <- function(rmst, sd){
diff <- diff(rmst)
diff_sd <- sqrt( sum(sd^2) )
p_val <- 2* (1 - pnorm( abs(diff/diff_sd) ))
c(diff = diff, sd = diff_sd, p = p_val)
}
x <- rbind( diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "sd"]),
diff_rmst(tmp$rmst[,"rmst"], tmp$rmst[, "wb_sd"]))
x <- as.data.frame(x)
x$lower <- x$diff - 1.96 * x$sd
x$upper <- x$diff + 1.96 * x$sd
round(x, digits = 3)
#> diff sd p lower upper
#> 1 0.883 0.396 0.026 0.106 1.660
#> 2 0.883 0.390 0.023 0.119 1.647Direct Estimator (Tian et.al 2014)
rmst2(time, status, group, tau= tau)
#>
#> The truncation time: tau = 24 was specified.
#>
#> Restricted Mean Survival Time (RMST) by arm
#> Est. se lower .95 upper .95
#> RMST (arm=1) 23.050 0.242 22.576 23.524
#> RMST (arm=0) 22.117 0.311 21.508 22.725
#>
#>
#> Restricted Mean Time Lost (RMTL) by arm
#> Est. se lower .95 upper .95
#> RMTL (arm=1) 0.950 0.242 0.476 1.424
#> RMTL (arm=0) 1.883 0.311 1.275 2.492
#>
#>
#> Between-group contrast
#> Est. lower .95 upper .95 p
#> RMST (arm=1)-(arm=0) 0.934 0.162 1.705 0.018
#> RMST (arm=1)/(arm=0) 1.042 1.007 1.079 0.018
#> RMTL (arm=1)/(arm=0) 0.504 0.278 0.914 0.024