Overview
metalite-table1.RmdMotivation
A table for descriptive statistics is widely used in medical research
and typically be the first table (i.e. table1) for a
manuscript.
There are R packages and examples to create the table1:
For use case in clinical trials, Chapter 4 of the R for clinical study reports and submission contains more details.
The metalite.table1 provide an interactive table1 to
enhance the communication between statisticians and clinicians.
Example
We use a subject level dataset r2rtf::r2rtf_adsl to
create a example interactive table1.
First, we modify the dataset by
- adding a few missing values.
- order the group variables.
- keep relevent variables.
# Prepare analysis ready data
df <- r2rtf_adsl
df$AGE[1:3] <- NA # Create missing value for illustration purpose.
df$ARM <- factor(
df$ARM,
c("Placebo", "Xanomeline Low Dose", "Xanomeline High Dose"),
c("Placebo", "Low Dose", "High Dose")
)
df <- df[, c("USUBJID", "ARM", "AGE", "SEX", "RACE", "BMIBLGR1")]
head(df)
#> USUBJID ARM AGE SEX RACE BMIBLGR1
#> 1 01-701-1015 Placebo NA F WHITE 25-<30
#> 2 01-701-1023 Placebo NA M WHITE >=30
#> 3 01-701-1028 High Dose NA M WHITE >=30
#> 4 01-701-1033 Low Dose 74 M WHITE 25-<30
#> 5 01-701-1034 High Dose 77 F WHITE 25-<30
#> 6 01-701-1047 Placebo 85 F WHITE >=30The interactive table1 can then be created by the
metalite_table1 function.
metalite_table1(
~ AGE + SEX + RACE + BMIBLGR1 | ARM, # formula
data = df, # source data
id = "USUBJID" # unique subject id
)The interactive features are illustrated in the GIF below.
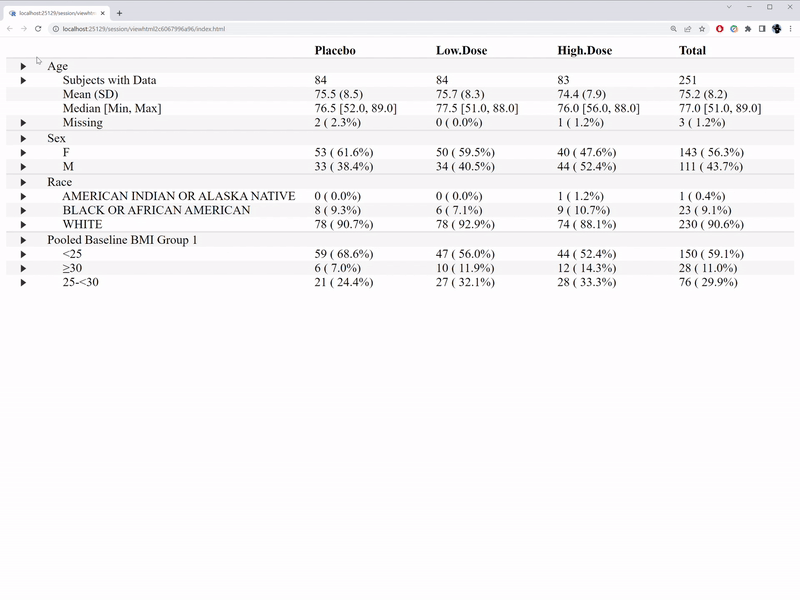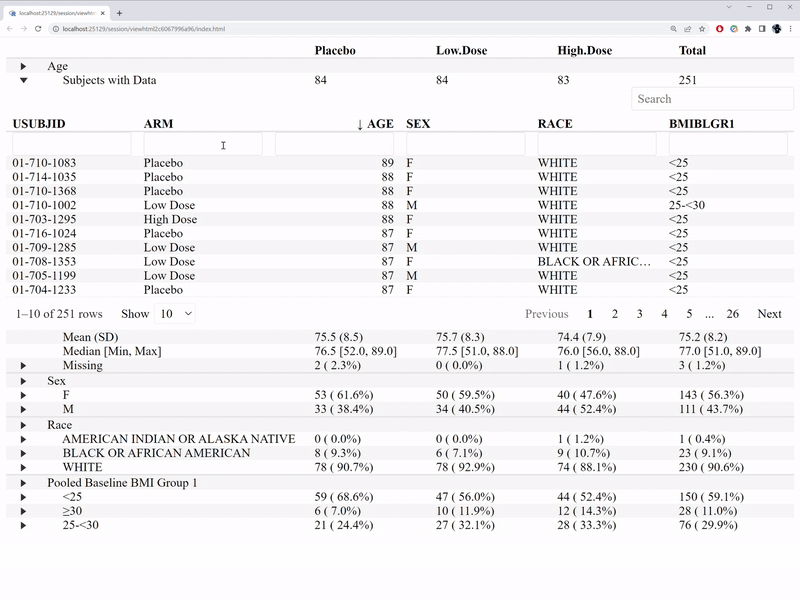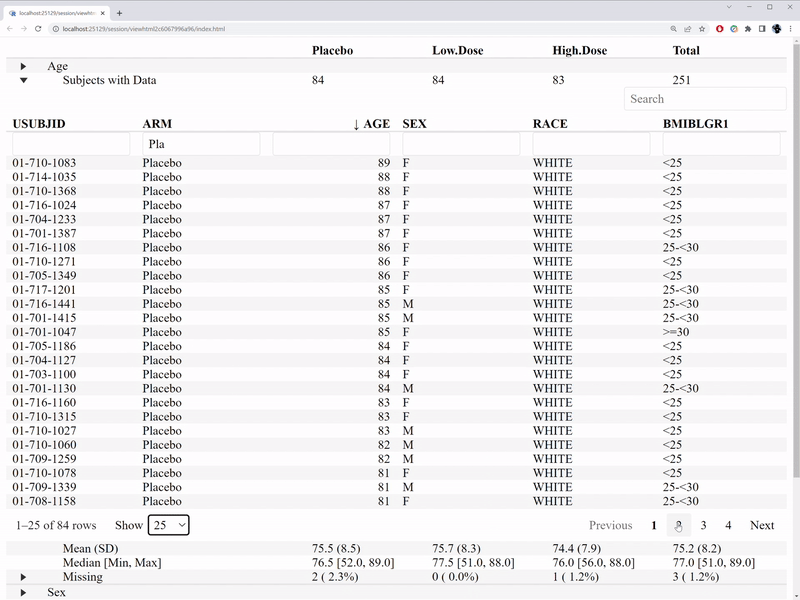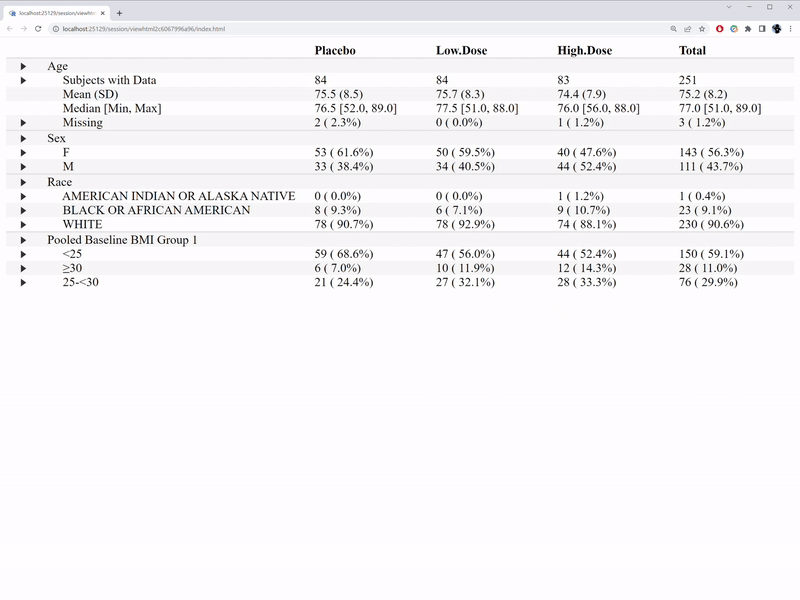
In comparison with the table1 R package, we are able to
answer ad-hoc questions from clinicians with interactive features such
as
- Who are those subjects with missing value.
- Who are those outliers.
- What’s the distribution of a variable by treatment group.
Those are common questions for ongoing clinical trials.
Features
Section Title
The section title is defined by the column labels. The
metalite::get_labels allows you quickly check the
label.
metalite::get_label(df)
#> USUBJID ARM
#> "Unique Subject Identifier" "ARM"
#> AGE SEX
#> "Age" "Sex"
#> RACE BMIBLGR1
#> "Race" "Pooled Baseline BMI Group 1"The metalite::assign_labels can modify labels
df <- metalite::assign_label(
df,
var = c("AGE", "BMIBLGR1"),
label = c("Age (Year)", "BMI Group (kg/m2)")
)
metalite::get_label(df)
#> USUBJID ARM
#> "Unique Subject Identifier" "ARM"
#> AGE SEX
#> "Age (Year)" "Sex"
#> RACE BMIBLGR1
#> "Race" "BMI Group (kg/m2)"The record_name argument can modify the term at the
first row, for example, “Number of Participants”.
metalite_table1(~ AGE + SEX | ARM,
data = df,
id = "USUBJID",
record_name = "Participants"
)Total Column
The Total column can be hided.
metalite_table1(~ AGE + SEX | ARM,
data = df,
id = "USUBJID",
total = FALSE
)Download Data
We can allow user to download raw data, for example we can set
download="listing" to enable user download the drill down
listing.
metalite_table1(~ AGE + SEX | ARM,
data = df,
id = "USUBJID",
download = "listing"
)Rmarkdown Render
Sometimes we want to create multiple tables in a for loop, we need to
clearly call the metalite_table1_to_html function and output
text as raw content by using resutls="asis" (e.g.,
{r, results="asis"}) in the Rmarkdown code chunk. Create a
dummy reactable object outside of for loop is necessary to
include all required javascript library. A full Rmarkdown example is as
below.
```{r}
library(metalite.table1)
```
```{r, include = FALSE}
reactable::reactable(data.frame(x = 1))
```
```{r, results="asis"}
type <- c("Subjects", "Records")
for (i in 1:2) {
cat("### Table ", i, "\n")
tbl <- metalite_table1(~ AGE + SEX | ARM,
data = r2rtf::r2rtf_adsl,
id = "USUBJID",
type = type[i]
)
metalite_table1_to_html(tbl)
}
```RTF file
User can also export a static table to an RTF file by using the
metalite_table1_to_rtf function.
metalite_table1(
~ AGE + SEX + RACE + BMIBLGR1 | ARM,
data = df,
id = "USUBJID"
) |>
metalite_table1_to_rtf("tmp1.rtf")
#> [1] "tmp1.rtf"